Tools and Systems
GIANT (giant.princeton.edu)
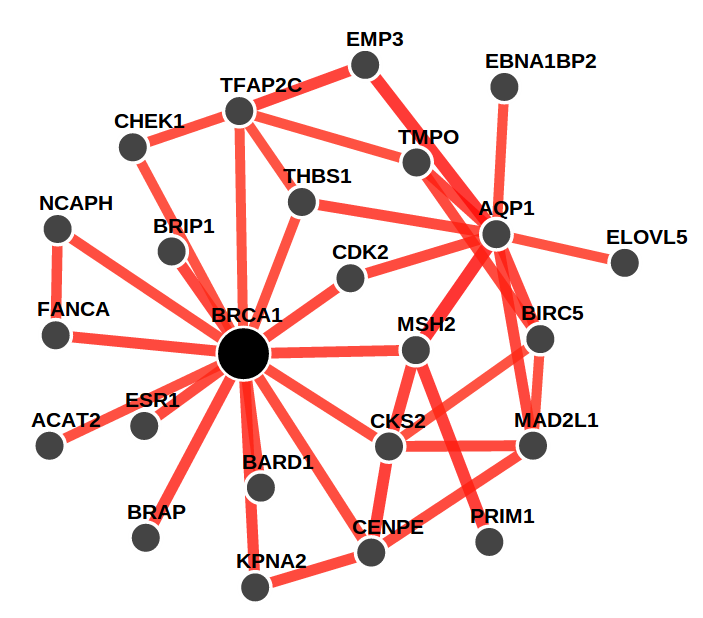
We present genome-wide functional interaction networks for 144 human tissues and cell types developed using a data-driven Bayesian methodology that integrates thousands of diverse experiments spanning tissue and disease states. Tissue-specific networks predict lineage-specific responses to perturbation, identify the changing functional roles of genes across tissues and illuminate relationships among diseases. We introduce NetWAS, which combines genes with nominally significant genome-wide association study (GWAS) P values and tissue-specific networks to identify disease-gene associations more accurately than GWAS alone. Our webserver, GIANT, provides an interface to human tissue networks through multi-gene queries, network visualization, analysis tools including NetWAS and downloadable networks. GIANT enables systematic exploration of the landscape of interacting genes that shape specialized cellular functions across more than a hundred human tissues and cell types. (giant.princeton.edu)
Curated Data Resource (cdr.pentaconhq.org)

PENTACON curators compiled lists of human genes involved in the Arachidonic Acid Pathway (AAP) and related networks based on the expertise of PENTACON researchers, as well as curated information from external resources including KEGG, Reactome, and GO. The genes were ranked as "Gold Standard", "Likely" or "Predicted" to be involved in the Arachidonic Acid or related pathways according to available experimental evidence. Specific gene curation was then performed by PENTACON curators based on information available in UniProt, BRENDA, BindingDB and the literature. The manually curated information, including chemicals, tissue specificity and disease involvement, was captured using standardized ontology terms. (cdr.pentaconhq.org)
AAP Pathway Viewer
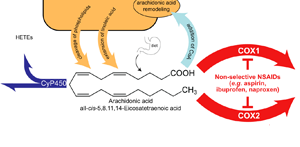
We created an interactive, graphical figure of the Arachidonic Acid Pathway to place the information we curated in its cellular context. The pathway figure is organized into four distinct quadrants showing the different origins and fates of arachidonic acid in the human body. When clicked, the graphical elements of the diagram display additional information and linkouts for each protein and eicosanoid in the pathway and for over 60 NSAIDs affecting the COX1 and/or COX2 enzymes. (AAP Pathway Viewer)
bii data repository (pentacon-bii.princeton.edu)

An extensive, annotated bii repository of large-scale data sets relevant to the NSAID response, including data sets published by the PENTACON consortium and by others. (pentacon-bii.princeton.edu)
Orthologs @CDR

